Programmable CRISPR-Based Epitranscriptomic Modifications for RNA Stability and Translation Control
Introduction to CRISPR Epitranscriptomics : RNA Stability and Translation Control
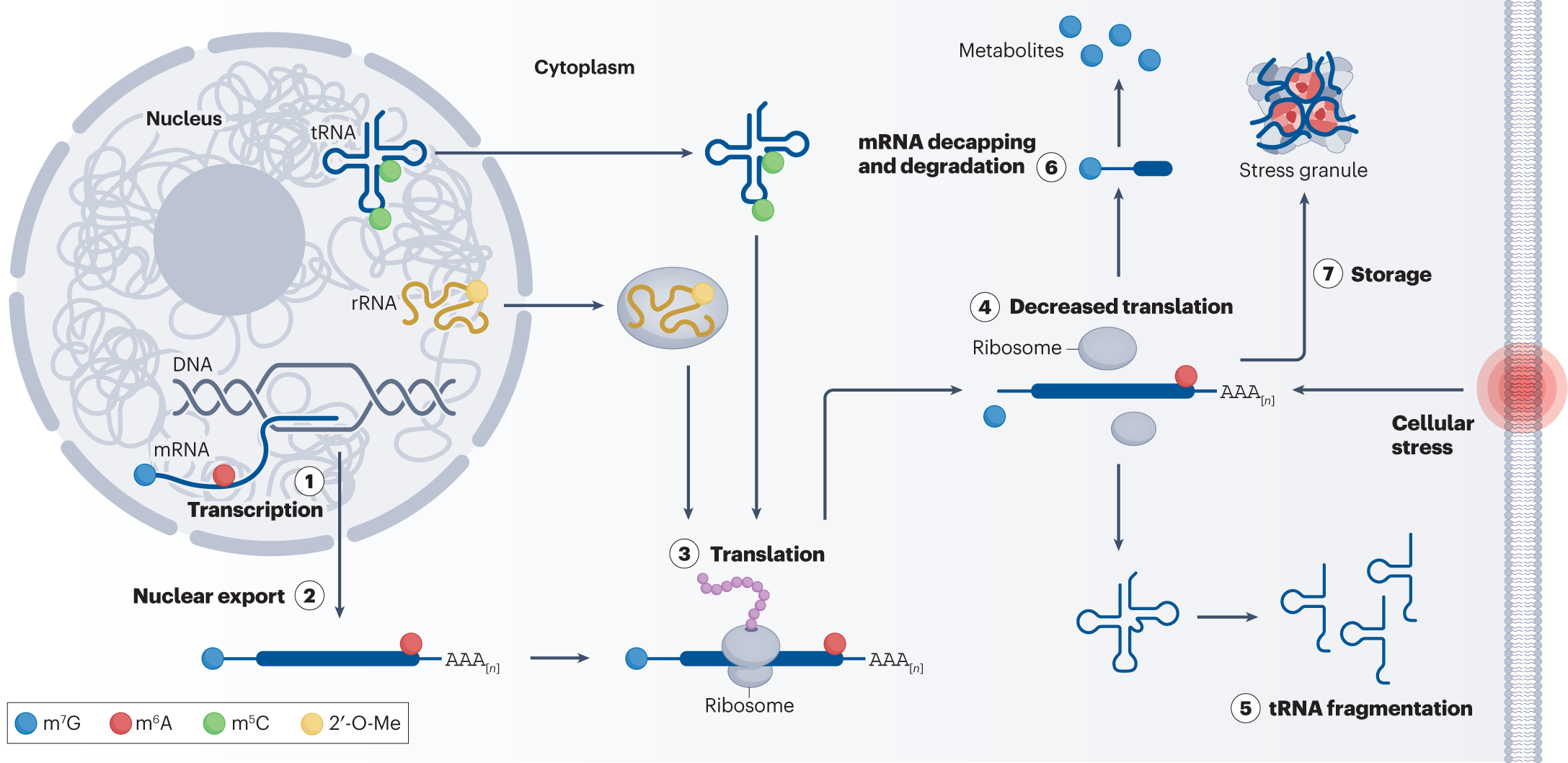
The CRISPR-Cas system has revolutionized gene editing, extending its capabilities from DNA modification to RNA-targeted regulation through epitranscriptomic modifications. Using programmable CRISPR-based RNA editing, researchers can influence mRNA stability, degradation pathways, and translation control, offering new approaches for post-transcriptional gene regulation (Abudayyeh et al., 2017).

CRISPR-Cas13 as a Programmable RNA Editing Tool
Unlike CRISPR-Cas9, which cuts DNA, CRISPR-Cas13 is designed for RNA targeting and modification. The catalytically inactive dCas13 variant can be fused with RNA-modifying enzymes to induce epitranscriptomic changes that impact RNA stability and translation efficiency (Cox et al., 2017). These modifications allow fine-tuning of mRNA turnover, ribosome binding, and translation regulation, making CRISPR-based RNA modifications a crucial tool for synthetic biology and molecular therapeutics.

Key Epitranscriptomic Modifications for RNA Stability and mRNA Regulation
1. m6A RNA Methylation for mRNA Stability
N6-methyladenosine (m6A) is one of the most abundant RNA modifications, influencing mRNA stability and decay. By recruiting m6A methyltransferases (e.g., METTL3/METTL14) via CRISPR-dCas13, scientists can enhance or suppress mRNA degradation pathways, impacting gene expression (Roundtree et al., 2017).

2. Adenosine-to-Inosine (A-to-I) RNA Editing for Post-Transcriptional Regulation
A-to-I RNA editing, catalyzed by adenosine deaminases acting on RNA (ADARs), alters mRNA sequences, modulating translation efficiency. CRISPR-Cas13 fused with ADARs enables programmable RNA modifications to modify codon usage and gene expression without altering DNA (Rees et al., 2017).

3. Pseudouridylation and RNA Stability
Pseudouridylation enhances mRNA stability and influences translation regulation. CRISPR-based RNA targeting combined with pseudouridine synthases can improve RNA processing and ribosome interaction, offering a promising approach for RNA therapeutics (Carlile et al., 2014).

CRISPR-Based Translation Control: Engineering mRNA Stability and Ribosome Binding
How Programmable RNA Modifications Regulate Translation
- mRNA Structure and Ribosome Interaction – Modified RNA stability elements affect ribosome binding and elongation rates.
- RNA-Targeted Codon Optimization – Programmable RNA modifications regulate translation efficiency at specific sites.
- Post-Transcriptional Gene Regulation – CRISPR-based RNA modifications influence protein synthesis dynamically.
Applications of CRISPR Epitranscriptomics in RNA Therapeutics and Gene Therapy
1. RNA-Based Therapeutics for Genetic Diseases
Programmable RNA editing via CRISPR-Cas13 can correct disease-associated RNA mutations, offering solutions for neurodegenerative disorders, cancer, and viral infections (Cox et al., 2017).

2. Synthetic Biology and Programmable Gene Circuits
CRISPR epitranscriptomics enables precision control over RNA stability and translation regulation, making it a key technology in synthetic biology for designing custom gene circuits and RNA-based drugs (Choudhury et al., 2020).

Future Perspectives: CRISPR-Based RNA Modifications in Medicine
The ability to precisely modulate RNA stability and translation control using CRISPR-based epitranscriptomics will drive advances in RNA-targeted therapeutics, post-transcriptional regulation, and synthetic biology applications.
=> CRISPR-based programmable RNA modifications provide an innovative approach to mRNA stability regulation and translation control. These tools will continue to shape RNA therapeutics, gene therapy, and molecular medicine, paving the way for next-generation RNA-based drug development.
References
- Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., ... & Zhang, F. (2017). RNA targeting with CRISPR–Cas13. Nature, 550(7675), 280-284.
- Carlile, T. M., Rojas-Duran, M. F., Zinshteyn, B., Shin, H., Bartoli, K. M., & Gilbert, W. V. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature, 515(7525), 143-146.
- Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., & Zhang, F. (2017). RNA editing with CRISPR-Cas13. Science, 358(6366), 1019-1027.
- Rees, H. A., & Liu, D. R. (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nature Reviews Genetics, 19(12), 770-788.
- Roundtree, I. A., Evans, M. E., Pan, T., & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169(7), 1187-1200.

