Unveiling the Structure-Function Relationship in Proteins: Why Shape Matters
Proteins are fundamental biomolecules that drive virtually every biological process within an organism. From catalyzing chemical reactions to providing structural support, the diverse roles of proteins depend heavily on their specific structures. The axiom "structure dictates function" applies most strongly in the field of protein biochemistry, where even a minor change in a protein’s conformation can have significant consequences on its function. Understanding the structure-function relationship in proteins provides critical insights into their biological roles, how mutations lead to diseases, and the design of targeted therapeutics.
Levels of Protein Structure: From Primary to Quaternary
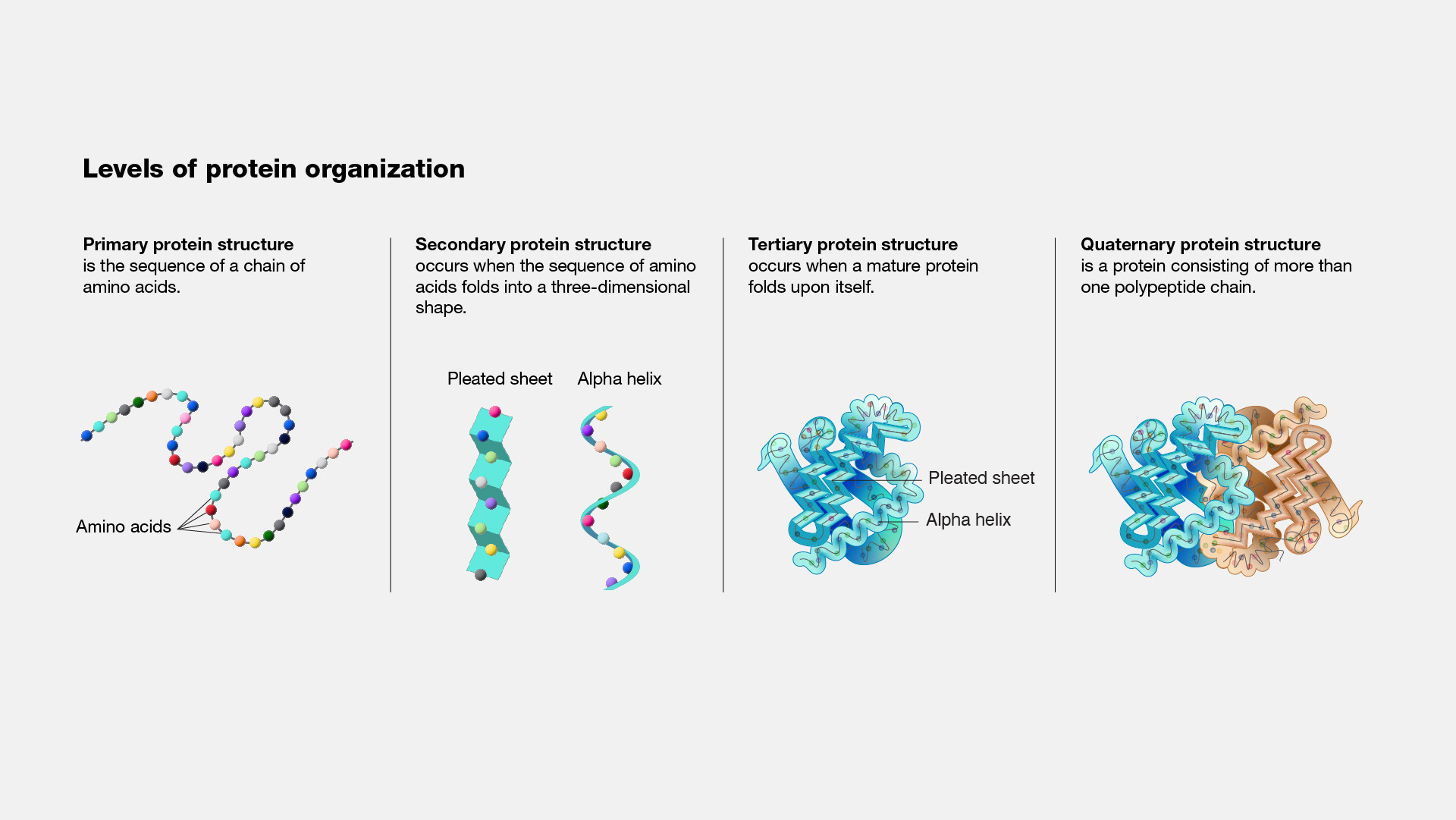
Proteins possess four levels of structural organization, each contributing to their final functional form.
- Primary Structure: This refers to the linear sequence of amino acids joined by peptide bonds. The sequence is determined by the gene encoding the protein, and even a single change in this sequence can alter the protein's overall structure and function. For example, in sickle cell anemia, a single amino acid substitution (valine for glutamic acid) in hemoglobin drastically changes its behavior, leading to disease.
- Secondary Structure: After the protein is synthesized, it begins to fold into regular structural motifs, primarily alpha-helices and beta-sheets. These structures are stabilized by hydrogen bonds between the backbone atoms of the peptide chain. Alpha-helices are coiled structures stabilized by intra-chain hydrogen bonding, while beta-sheets are formed by inter-strand hydrogen bonding, which can be parallel or antiparallel. These secondary structures form the scaffold on which the protein's tertiary structure is built.
- Tertiary Structure: The tertiary structure refers to the three-dimensional arrangement of all atoms within a single polypeptide chain, including the folding of alpha-helices and beta-sheets into compact domains. Tertiary structure is stabilized by a range of interactions, including hydrophobic interactions, disulfide bridges, hydrogen bonds, and ionic bonds between side chains. This level of structure is essential for the protein's specific function. For example, enzymes require a precise arrangement of residues within their active sites to catalyze reactions effectively.
- Quaternary Structure: Some proteins function as complexes composed of multiple polypeptide chains, or subunits. The quaternary structure refers to how these subunits interact and assemble. Hemoglobin, for example, consists of four subunits (two alpha and two beta chains), and this quaternary structure is crucial for its ability to bind and transport oxygen efficiently.
The Role of Protein Folding in Function
Protein folding is the process by which a polypeptide chain attains its functional, three-dimensional structure. This folding is not a random process; rather, it is directed by the sequence of amino acids in the chain (primary structure), which dictates the eventual secondary, tertiary, and quaternary structures. The folding process is energetically driven to achieve the most thermodynamically stable configuration. Misfolded proteins, however, can lead to dysfunction, aggregation, and disease.
- Chaperones and Assisted Folding: Cells use molecular chaperones to assist in the folding of newly synthesized proteins and to prevent misfolded proteins from aggregating. Chaperones do not determine the final structure but rather facilitate the correct folding process by providing a suitable environment.
- Protein Misfolding and Disease: When proteins fail to fold properly, they can aggregate into insoluble fibrils or plaques, leading to diseases such as Alzheimer's and Parkinson's. In Alzheimer's disease, for instance, the misfolding of amyloid-beta peptides leads to plaque formation, which disrupts neural function. The structure-function relationship here is critical, as a misfolded protein loses its normal function and often gains toxic properties.
Protein Domains: Modular Units of Function
Many proteins are composed of distinct regions known as domains. Each domain is a functional unit that can fold independently and often carries out a specific task. For example, enzymes may have separate domains for substrate binding and catalysis. The modular nature of protein domains allows for versatility in protein function, as domains can be rearranged or combined to generate proteins with new or enhanced functions.
- SH2 and SH3 Domains in Signaling Proteins: SH2 (Src homology 2) and SH3 (Src homology 3) domains are common motifs in proteins involved in signal transduction. SH2 domains bind to phosphorylated tyrosine residues, while SH3 domains recognize proline-rich regions. Together, these domains enable proteins to recognize and bind to specific partners in signaling pathways, controlling key cellular processes such as growth and differentiation.
Allosteric Regulation: Shape Changes and Functional Control
One of the most intriguing aspects of protein structure is its dynamic nature. Many proteins undergo conformational changes in response to ligand binding or other environmental stimuli. This property, known as allosteric regulation, is central to the function of many enzymes and receptors.
- Hemoglobin and Oxygen Binding: Hemoglobin is a classic example of an allosteric protein. In the lungs, hemoglobin binds oxygen, causing a conformational change that increases its affinity for additional oxygen molecules. In the tissues, where oxygen levels are low, hemoglobin releases oxygen, undergoing another conformational shift. This cooperative binding and release of oxygen are crucial for efficient oxygen transport in the body.
- Enzymes and Feedback Inhibition: Allosteric regulation also plays a significant role in enzyme activity. Certain enzymes have allosteric sites where the binding of a regulatory molecule induces a conformational change, either activating or inhibiting the enzyme’s catalytic activity. This mechanism is often seen in metabolic pathways, where the end product of the pathway can inhibit an enzyme at the beginning of the pathway, preventing overproduction.
Structural Flexibility and Protein Function
While many proteins adopt a single, stable structure, others exhibit structural flexibility that is integral to their function. Intrinsically disordered proteins (IDPs) or regions within proteins do not fold into a fixed three-dimensional structure. Instead, they remain flexible and dynamic, allowing them to participate in multiple interactions and functions.
- p53: A Flexible Guardian of the Genome: The tumor suppressor protein p53 is an example of a protein with both structured and intrinsically disordered regions. p53 plays a critical role in regulating the cell cycle and preventing cancer. Its disordered regions allow it to interact with a wide variety of other proteins and regulatory molecules, while its structured domains are responsible for specific functions, such as DNA binding.
Protein Engineering and Structure-Based Drug Design
The understanding of protein structure-function relationships has opened the door to protein engineering, where scientists design proteins with novel functions or improved properties. This is particularly useful in the development of biopharmaceuticals and industrial enzymes.
- Rational Design and Directed Evolution: In rational design, researchers modify specific amino acids in a protein based on knowledge of its structure to enhance or alter its activity. Directed evolution, on the other hand, mimics natural selection by generating a library of protein variants and selecting for those with desired traits. These approaches have led to the development of proteins with improved stability, activity, or substrate specificity.
- Structure-Based Drug Design: In drug discovery, the knowledge of protein structures enables the design of molecules that can specifically bind to a protein’s active or regulatory site, modulating its function. This is the basis of many modern therapies, including inhibitors of enzymes like proteases in viral infections or kinases in cancer.
The relationship between protein structure and function is fundamental to all aspects of biology. Understanding how the intricate architecture of proteins dictates their roles in the cell allows us to comprehend how life works at the molecular level. It also enables us to devise strategies for treating diseases caused by structural malfunctions, develop engineered proteins for industrial applications, and design drugs that specifically target disease-related proteins. As our ability to visualize and manipulate protein structures improves, the opportunities for harnessing this knowledge will continue to expand.
Recent Posts
-
Can mNGS Replace Culture?
In microbiology and infectious-disease work, culture has been the gold standard for over a century. …30th Sep 2025 -
Post-Translational Modifications: The Hidden Layers of Protein Regulation
Proteins are the workhorses of cellular biology, performing a wide array of functions that are essen …18th Oct 2024 -
Unveiling the Structure-Function Relationship in Proteins: Why Shape Matters
Proteins are fundamental biomolecules that drive virtually every biological process within an organi …18th Oct 2024