Post-Translational Modifications: The Hidden Layers of Protein Regulation
Proteins are the workhorses of cellular biology, performing a wide array of functions that are essential for life. While the genetic code determines the primary amino acid sequence of proteins, their final functionality is often modulated by post-translational modifications (PTMs). These modifications, which occur after protein synthesis, can alter the chemical properties, localization, activity, and interaction patterns of proteins. PTMs introduce a hidden layer of regulation, adding versatility and complexity to protein function that goes beyond what is encoded in the DNA.
In this blog, we will explore the various types of post-translational modifications, their mechanisms, and the crucial roles they play in cellular processes. The technical focus will shed light on the biochemical underpinnings of these modifications and how they integrate into broader biological systems.
What Are Post-Translational Modifications (PTMs)?
Post-translational modifications refer to the covalent processing of proteins after they are translated from mRNA on ribosomes. These modifications involve the addition or removal of specific chemical groups, proteins, lipids, or carbohydrates, and are often enzymatically catalyzed. PTMs can fine-tune the function of a protein by altering its physical and chemical properties, including its stability, charge, localization, and interaction with other biomolecules.
Common types of PTMs include:
- Phosphorylation
- Ubiquitination
- Acetylation
- Methylation
- Glycosylation
- Sumoylation
- Lipidation
- Proteolytic cleavage
Each of these modifications serves as a distinct regulatory switch, turning protein functions on or off, or modifying them in more nuanced ways. The interplay of multiple PTMs on a single protein can create a dynamic regulatory network known as the "PTM code."
Phosphorylation: A Key Regulator of Protein Activity
Phosphorylation is one of the most common and well-studied post-translational modifications. It involves the addition of a phosphate group (PO₄³⁻) to the hydroxyl group of specific amino acids—typically serine, threonine, or tyrosine—within a protein. The modification is catalyzed by enzymes known as protein kinases, while phosphatases remove these groups.
Phosphorylation serves as a molecular switch that can activate or deactivate enzymes, alter protein conformation, and regulate interactions between proteins. It is central to numerous signaling pathways, especially those involved in cellular growth, metabolism, and apoptosis.
- Mechanism: Phosphorylation introduces a negatively charged phosphate group, which can alter the protein's overall charge distribution, leading to conformational changes. This change may either expose or occlude active sites or binding regions, thereby modulating the protein’s function.
- Example: In the mitogen-activated protein kinase (MAPK) signaling pathway, a cascade of phosphorylation events transmits signals from the cell surface to the nucleus. Here, phosphorylation of kinases like ERK (extracellular signal-regulated kinase) results in its activation, enabling it to phosphorylate downstream transcription factors that regulate gene expression.
Phosphorylation also plays a critical role in cell cycle regulation. Cyclin-dependent kinases (CDKs) rely on phosphorylation events to transition the cell through different phases of the cell cycle, ensuring proper division and replication.
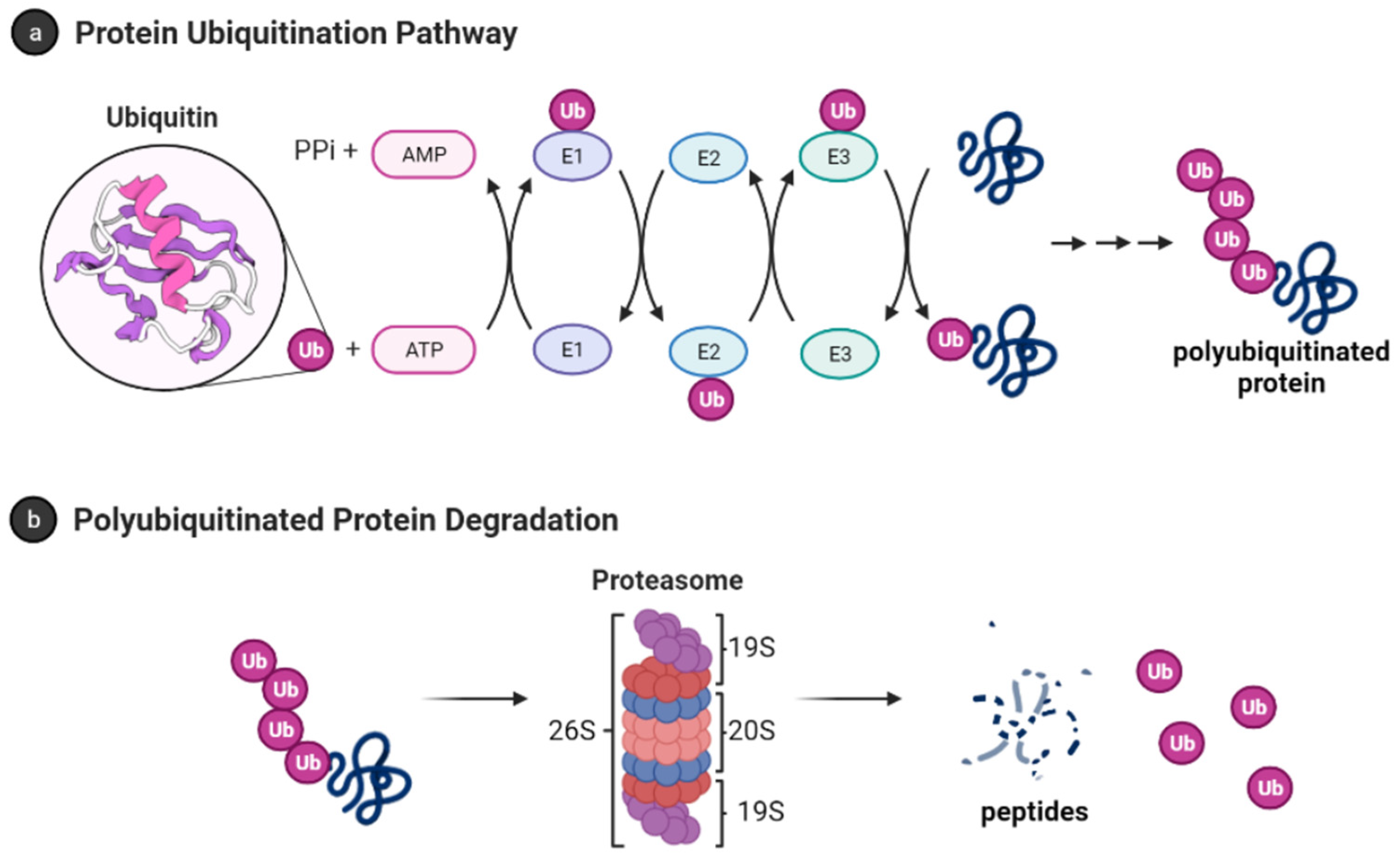
Ubiquitination: The Tagging System for Protein Degradation
Ubiquitination is a process where a small protein called ubiquitin is covalently attached to lysine residues on target proteins. This modification serves as a signal for protein degradation via the proteasome, a large proteolytic complex. Ubiquitination also has non-degradative roles, such as regulating protein localization, signal transduction, and DNA repair.
- Mechanism: Ubiquitination occurs through a cascade involving three types of enzymes:
- E1 (Ubiquitin-activating enzyme): Activates ubiquitin in an ATP-dependent manner.
- E2 (Ubiquitin-conjugating enzyme): Transfers the activated ubiquitin to the substrate.
- E3 (Ubiquitin ligase): Confers substrate specificity by catalyzing the transfer of ubiquitin from the E2 enzyme to the lysine residue of the target protein.
- Types of Ubiquitination:
- Monoubiquitination: The addition of a single ubiquitin molecule to a substrate, which often regulates processes such as endocytosis or DNA repair.
- Polyubiquitination: The attachment of a ubiquitin chain, where ubiquitins are linked to one another. This typically serves as a signal for proteasomal degradation (especially when ubiquitin chains are linked via lysine 48).
- Example: The tumor suppressor p53 is regulated by ubiquitination. Under normal conditions, the E3 ligase MDM2 ubiquitinates p53, leading to its degradation. In response to DNA damage, this ubiquitination is inhibited, allowing p53 to accumulate and trigger cell cycle arrest or apoptosis.
Acetylation and Methylation: Fine-Tuning Gene Expression
Both acetylation and methylation are critical PTMs that regulate chromatin structure and gene expression. These modifications occur predominantly on histone proteins but can also modify non-histone proteins, influencing a wide range of cellular functions.
- Acetylation: Involves the addition of an acetyl group (CH₃CO) to lysine residues, primarily on histones. Histone acetylation neutralizes the positive charge on lysine, reducing its interaction with the negatively charged DNA, leading to a more relaxed chromatin structure and increased transcriptional activity.
- Example: Acetylation of histones by histone acetyltransferases (HATs) generally promotes gene expression. Conversely, histone deacetylases (HDACs) remove these acetyl groups, compacting chromatin and silencing genes.
- Methylation: Involves the addition of methyl groups (CH₃) to lysine or arginine residues. Depending on the site and number of methyl groups added (mono-, di-, or trimethylation), methylation can either activate or repress transcription.
- Example: Trimethylation of histone H3 at lysine 4 (H3K4me3) is associated with active transcription, whereas trimethylation at lysine 9 (H3K9me3) is associated with gene silencing.
Acetylation and methylation of non-histone proteins can also affect protein stability, localization, and interactions.
Glycosylation: The Addition of Sugar Moieties
Glycosylation is the covalent attachment of carbohydrate groups to asparagine (N-linked glycosylation) or serine/threonine (O-linked glycosylation) residues. This modification is essential for the proper folding, stability, and function of many proteins, particularly those secreted or located on the cell surface.
- N-linked Glycosylation: Occurs in the endoplasmic reticulum (ER) and involves the attachment of a preassembled oligosaccharide to asparagine residues within the consensus sequence Asn-X-Ser/Thr.
- O-linked Glycosylation: Takes place in the Golgi apparatus, where sugar moieties are added sequentially to serine or threonine residues.
Glycosylation influences protein folding by stabilizing intermediate conformations and protecting proteins from degradation. It also plays a critical role in cell-cell communication, immune responses, and pathogen recognition.
- Example: Glycoproteins on the surface of immune cells contain specific glycan structures that are recognized by lectins on other cells. This interaction is critical for immune surveillance and the response to infections.
Sumoylation: A Modifier of Nuclear and Cytoskeletal Functions
Sumoylation is the attachment of a small ubiquitin-like modifier (SUMO) protein to lysine residues on target proteins. Unlike ubiquitination, sumoylation typically does not signal for protein degradation but rather modulates protein localization, stability, and interaction networks.
- Mechanism: Sumoylation is catalyzed by a similar cascade of enzymes (E1, E2, and E3) as ubiquitination but involves the SUMO protein.
- Example: Sumoylation of transcription factors can alter their ability to bind DNA or recruit co-factors, thereby regulating gene expression. One key example is the sumoylation of the transcription factor STAT1, which regulates its activity in response to interferon signaling.
Lipidation: Membrane Targeting and Anchoring
Lipidation involves the covalent attachment of lipid groups to proteins, facilitating their association with cellular membranes. Common forms of lipidation include myristoylation, palmitoylation, and prenylation.
- Myristoylation: The attachment of a myristoyl group (derived from myristic acid) to the N-terminal glycine of a protein. This modification typically targets proteins to the inner leaflet of the plasma membrane.
- Example: Src-family kinases undergo myristoylation, allowing them to associate with membranes where they phosphorylate downstream signaling proteins.
- Palmitoylation: Involves the attachment of palmitic acid to cysteine residues, further stabilizing protein-membrane interactions.
- Prenylation: The addition of isoprenoid groups to cysteine residues near the C-terminus, targeting proteins to the membrane.
Lipidation is essential for the localization and function of many signaling proteins, including G-proteins and Ras.
Post-translational modifications provide an additional layer of regulation that enables proteins to perform diverse and complex functions. Through PTMs, proteins can be dynamically regulated in response to environmental cues, ensuring proper cellular function. Understanding the mechanisms and roles of these modifications offers crucial insights into health, disease, and therapeutic interventions. As proteomics technologies advance, uncovering more PTMs and their interplay will further illuminate the intricate regulatory networks that govern cellular biology.
Recent Posts
-
Can mNGS Replace Culture?
In microbiology and infectious-disease work, culture has been the gold standard for over a century. …30th Sep 2025 -
Post-Translational Modifications: The Hidden Layers of Protein Regulation
Proteins are the workhorses of cellular biology, performing a wide array of functions that are essen …18th Oct 2024 -
Unveiling the Structure-Function Relationship in Proteins: Why Shape Matters
Proteins are fundamental biomolecules that drive virtually every biological process within an organi …18th Oct 2024